Deploy your R models
Welcome to jaqpotr documentation
jaqpotr is a library that lets you deploy and share machine learning and statistal models developed in R (https://www.r-project.org/) over the web as ready to use web services.
Installation
jaqpotr can be installed directly from GitHub using the install_github function of the devtools package.
devtools::install_github("euclia/jaqpotr")
Logging in using Jaqpot credentials
If you have created a Jaqpot account, you can log into Jaqpot using your Jaqpot credentials with login.cred() . Once executed, a prompt message pops up asking for your Jaqpot username, followed by a pop-up window in which you should fill in your Jaqpot password.
Logging in using API key
If you want to log in using an API key, then use login.api(). Once executed, a pop-up window appears in which you should fill in your Jaqpot password.
The API key can be downloaded as a text file from the Jaqpot User Interface.
Usage
Now all you need to do is call the desired jaqpotr function with the appropriate input arguments. Once you log in, use each deployment function appropriately. Once executed, the only outstanding step is to provide a title and a short description for your model. Be wary of the fact that the model title cannot change, whereas the model description can be accessed and further processed via the Jaqpot User Interface. Upon successful deployment on Jaqpot, your model, which is now effeectively a web service, is given a unique model ID!
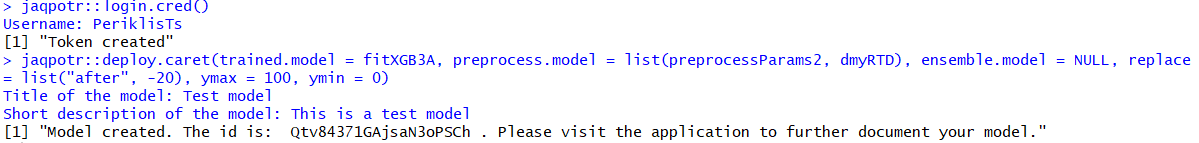
Your model is then available at https://app.jaqpot.org
For some R models, your data might be uploaded in an encrypted form on Jaqpot. We are currently working on resolving this, so that all data remain on the local system and only the necessary model parameters and metadata arre uploaded on Jaqpot.
Supported R packages
At the moment Jaqpot can host models developed is specific R libraries. The available jaqpotr functions are:
deploy.pbpk()deploys an ODE model on Jaqpot that can be solved using theodefunction of the deSolve packagedeploy.gbm()deploys an object of class "gbm" developed using thegbm()function of the gbm packagedeploy.lm()deploys an object of class "lm" or "glm" developed using thelm()orglm()functions of the base packagedeploy.nb ()deploys an object of class "naive_bayes" developed using thenaive_bayes()function of the naivebayes packagedeploy.randomForest()deploys an object of class "randomForest.formula" developed using therandomForest()function of the randomForest packagedeploy.rpart()deploys an object of class "rpart" developed using therpart()function of the rpart packagedeploy.svm()deploys an object of class "svm.formula" developed using thesvm()function of the e1071 packagedeploy.tree()deploys an object of class "tree " developed using thetree()function of the tree packagedeploy.bn()deploys an object of class "bn.fit " developed using thebn.fit()function of the bnlearn packagedeploy.caret()deploys objects developed using the caret package with one of the following methods:"xgbTree""neuralnet""rf""svmLinear2""gbm"
If you want to deploy a model that has been developed in a library that is not part of the list, all you need to do is contact us and we will develop the appropriate function as soon as possible!
Example 1: Deploy a simple linear regression model
library(jaqpotr)
library(mlbench) # contains the BostonHousing dataset
data(BostonHousing)
lm.model <- lm(medv ~ ., data=BostonHousing)
deploy.lm(lm.model)
The names of the dependent and independent features that will appear on the Jaqpot User Interface of the model will be the same as the ones provided to the model during the training phase and can't change. Therefore, carefully name your variables before training the model. In the example above, the input varialbes will be named according to the BostonHousing dataframe column names, and the prediction will be named 'medv'.
Example 2: Deploy a naive bayes model
library(naivebayes)
library(jaqpotr)
data(iris)
nb.model <- naive_bayes(Species ~., data = iris)
deploy.nb(nb.model)
Example 3: Include input preprocessing using deploy.caret
deploy.caret() offers the chance to incorporate preprocessing pipeliness implemented in caret, through an extra argument, preprocess.model, which defaults to NULL. In particular, a series of preprocessing caret functions can be applied to the user input, before it is fed to the trained model object. For achieving this, the only thing you have to do is to provide a list with all the preprocessing models you used.
library(jaqpotr)
library(caret) # Needed for preprocessing and model training
library(xgboost) #Needed for building the xgb model
library(mlbench) #Contains the BostonHousing dataset
data(BostonHousing)
X_train <- data.frame(BostonHousing[, -which(names(BostonHousing) == "medv")])
Y_train <- data.frame(medv = (BostonHousing[, "medv"] )) #slicing omits the variabe name, so use dataframe and retain it
# Preprocessing
# Step 1: Bring all independent values in the [0,1] interval
preprocessParams <- preProcess(X_train, method="range")
X_train <- predict(preprocessParams, X_train)
# Step 2: One hot encoding applied to categorical variables
dmyRDT <- dummyVars(~.,data=X_train)
X_train <- predict(dmyRDT, newdata=X_train)
train_df <- cbind(X_train, Y_train)
## Build the model
trainControl <- trainControl(method="repeatedcv", number=20, repeats=3,verboseIter = FALSE,allowParallel = FALSE, savePredictions=TRUE)
tunegrid <- expand.grid(nrounds = 100,
max_depth = 2,
eta = 0.20,
gamma =0 ,
colsample_bytree =0.6 ,
min_child_weight =1,
subsample =1
)
fitXGB2 <- train(medv ~ ., data = train_df, method="xgbTree", trControl=trainControl ,tuneGrid=tunegrid,importance=TRUE,savePredictions = 'final')
# Deploy the model on Japot
deploy.caret(trained.model = fitXGB2, preprocess.model = list(preprocessParams,dmyRDT))
The preprocess models must include only the independent variables, not the dependent!! Additionally, you must provide the preprocess models in the order applied, in a list form.
Example 4: Deploy a custom PBK model on Jaqpot
deploy.pbpk() offers the chance to upload custom R PBK and other biokinetics models that are expressed through differntial equations. The user should provide the inputs that the end-user will provide on the Jaqpot User Interface (UI) and the state variables of the ODE system that are to be printed on the UI upon execution of the ODE system. In addition, the user should provide five functions. The first function transforms the user input according to the needs of the ODE model, the
second creates the initial conditions of the ODEs, the third creates the events that are forced on the system, the fourth enables the use of custom functions inside the ODEs and the last is the ODEs, with syntax compatible with package deSolve. The functions are used in a nested style (see the following example). Note that the names of independent and dependent features (i.e. user.input) cannot be further modified via the Jaqpot UI, so the user should choose them with caution.
user.input <-list("weight" = 250,"dose" = c(10,12), "administration.time" = c(0,1.5) )
out.vars <- c("Li")
##########################################
# Function for creating parameter vector #
##########################################
create.params <- function(input){
with( as.list(input),{
############################
# Physiological parameters #
############################
# tissue weights (in g)
W_tot <- weight # ;body weight, experimental data - g
W_lu <-1.2 # weight of lungs, experimental data - g
W_li <- 10.03 # weight of liver, experimental data - g
W_blood <- 0.065 * W_tot
W_rob <- W_tot - (W_blood + W_li + W_lu)
#Regional blood flows (in mL per hour)
fQl = 0.183 # fraction of cardiac output to liver, unitless
fQrob = 1-fQl # fraction of cardiac output to rest of the body, unitless
Q_tot <- 4980 # cardiac output, mL/h
Q_li <- fQl*Q_tot # blood flow to liver, mL/h
Q_rob <- fQrob*Q_tot # blood flow to rest of the body, mL/h
P <-1.435445 # partition coefficient tissue:blood, unitless
CLE_f <- 9.958839e-05 # clearance rate to feces from liver, mL/h
return(list("W_lu" = W_lu, "W_li" = W_li, "W_rob" = W_rob, "W_blood" = W_blood, "Q_tot" = Q_tot,
"Q_li" = Q_li, "Q_rob" = Q_rob, "P" = P,"CLE_f" = CLE_f, "dose" = dose))
})
}
### store the values
params <- create.params(user_input)
#################################################
# Function for creating initial values for ODEs #
#################################################
create.inits <- function(parameters){
with( as.list(parameters),{
Lu <- 0; Rob <- 0;Li <- 0; Art_blood <- 0; Ven_blood <- 0;
return(c("Lu" = Lu, "Rob" = Rob, "Li" = Li, "Art_blood" = Art_blood,
"Ven_blood" = Ven_blood))
})
}
##store the values
inits <- create.inits(params)
#################################################
# Function for creating events #
#################################################
create.events<- function(parameters){
with( as.list(parameters),{
ldose <- length(dose)
ltimes <- length(administration.time)
addition <- dose
if (ltimes == ldose){
events <- list(data = rbind(data.frame(var = "Ven_blood", time = administration.time,
value = addition, method = c("add")) ))
}else{
stop("The times when the drug is injected should be equal in number to the doses")
}
return(events)
})
}
events <- create.events(params)
###################
# Custom function #
###################
custom.func <- function(W_li){
if (W_li<15){
a = 10
}else{
a = 15
}
return(a)
}
#################
# ODEs system #
#################
ode.fun <- function(time, Initial.values, Parameters, custom.func){
with( as.list(c(Initial.values, Parameters)),{
#cleararance coefficient
cl = custom.func(weight)
# concentrations in tissues
C_lu <- Lu/W_lu
C_re <- Rob/W_re
C_li <- Li_tissue/W_li
C_art <- Art_blood/(0.2*W_blood)
C_ven <- Ven_blood/(0.8*W_blood)
# Lungs
dlu <- Q_tot * (C_ven - C_lu/P)
# Rest of the body
dRob_tissue <- Q_rob * (C_art - C_rob/P)
# Liver
dLi_tissue <- Q_li * (C_art - C_li/P)- CLE*cl*C_li
# Arterial blood
dArt_blood <- Q_tot* C_lu/P - Cart * (Q_li + Q_rob)
# Venous blood
dVen_blood <- Q_li*C_li/P + Q_rob*C_rob/P - Q_tot * C_ven
list(c(dLu = dLu, dRob = dRob, dLi = dLi, dArt_blood = dArt_blood, dVen_blood = dVen_blood)
})
}
deploy.ode(user.input, out.vars, create.params, create.inits, create.events, custom.func, ode.fun, method = "bdf", list(rtol=1e-07, atol=1e-09)
}
About
The jaqpotr library has been developed by the Unit of Process Control and Informatics in the School of Chemical Engineering at the National Technical University of Athens and is supported by Euclia.